
All the sessions from Transform 2021 are accessible on-demand now. Watch now.
DeepMind and the European Bioinformatics Institute (EMBL), a life sciences lab based in Hinxton, England, today announced the launch of what they claim is the most full and correct database of structures for proteins expressed by the human genome. In a joint press conference hosted by the journal Nature, the two organizations mentioned that the database, the AlphaFold Protein Structure Database, which was made applying DeepMind’s AlphaFold 2 method, will be made accessible to the scientific neighborhood in the coming weeks.
The recipe for proteins — significant molecules consisting of amino acids that are the basic constructing blocks of tissues, muscle tissues, hair, enzymes, antibodies, and other vital components of living organisms — are encoded in DNA. It’s these genetic definitions that circumscribe their 3-dimensional structures, which in turn figure out their capabilities. But protein “folding,” as it is named, is notoriously complicated to figure out from a corresponding genetic sequence alone. DNA consists of only details about chains of amino acid residues and not these chains’ final kind.
Image Credit: DeepMind
In December 2018, DeepMind attempted to tackle the challenge of protein folding with AlphaFold, the item of two years of work. Its successor, AlphaFold 2, announced in December 2020, enhanced on this to outgun competing protein-folding-predicting techniques. In the benefits from the 14th Critical Assessment of Structure Prediction (CASP) assessment, AlphaFold 2 had typical errors comparable to the width of an atom (or .1 of a nanometer), competitive with the benefits from experimental techniques.
“The AlphaFold database shows the potential for AI to profoundly accelerate scientific progress. Not only has DeepMind’s machine learning system greatly expanded our accumulated knowledge of protein structures and the human proteome overnight, its deep insights into the building blocks of life hold extraordinary promise for the future of scientific discovery,” Alphabet and Google CEO Sundar Pichai mentioned in a press release.
Illuminating protein structures
AlphaFold 2 draws inspiration from the fields of biology, physics, and machine finding out, taking benefit of the truth that a folded protein can be believed of as a “spatial graph” exactly where amino acid residues (amino acids contained inside a peptide or protein) are nodes, and edges connect the residues in close proximity. AlphaFold 2 leverages an AI algorithm that attempts to interpret the structure of this graph although reasoning more than the implicit graph it is constructing, applying evolutionarily associated sequences, numerous sequence alignment, and a representation of amino acid residue pairs.
In an open supply codebase published last week, DeepMind drastically streamlined AlphaFold 2. Whereas the close-sourced method took days of computing time to produce structures, the open supply version is about 16 instances quicker and can create structures in minutes to hours, based on the protein size.
These improvements enabled DeepMind and the EMBL to develop more than than 350,000 protein structure predictions such as the human proteome (which spans 20,000 proteins), more than doubling the quantity of higher-accuracy structures accessible to researchers. Beyond this, DeepMind and EMBL utilised AlphaFold 2 to predict the structures of 20 other “biologically significant organisms,” yielding more than 350,000 structures in total for E. coli, fruit flies, mice, zebrafish, yeast, malaria parasites, tuberculosis bacteria, and more. The program is to expand coverage to more than one hundred million structures as improvements to each AlphaFold 2 and the database come on line.
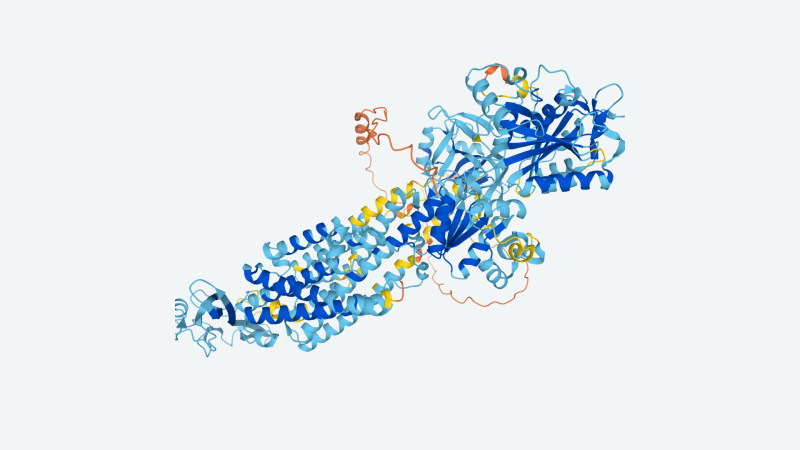
Image Credit: DeepMind
“This will be one of the most important datasets since the mapping of the Human Genome,” EMBL deputy director common Ewan Birney mentioned in a statement. “Making AlphaFold 2 predictions accessible to the international scientific community opens up so many new research avenues, from neglected diseases to new enzymes for biotechnology and everything in between. This is a great new scientific tool, which complements existing technologies, and will allow us to push the boundaries of our understanding of the world.”
Some scientists caution that AlphaFold 2 is not probably the finish-all be-all when it comes to protein structure prediction. Steven Finkbeiner, professor of neurology at the University of California, San Francisco, told Wired in an interview that it is as well quickly to inform the implications for drug discovery, offered the wide variation in structures inside the human body. But DeepMind tends to make the case that AlphaFold 2, if additional refined, could be applied to previously intractable difficulties, such as these associated to epidemiological efforts. Last year, the organization predicted many protein structures of SARS-CoV-2, such as ORF3a, whose makeup was formerly a mystery.
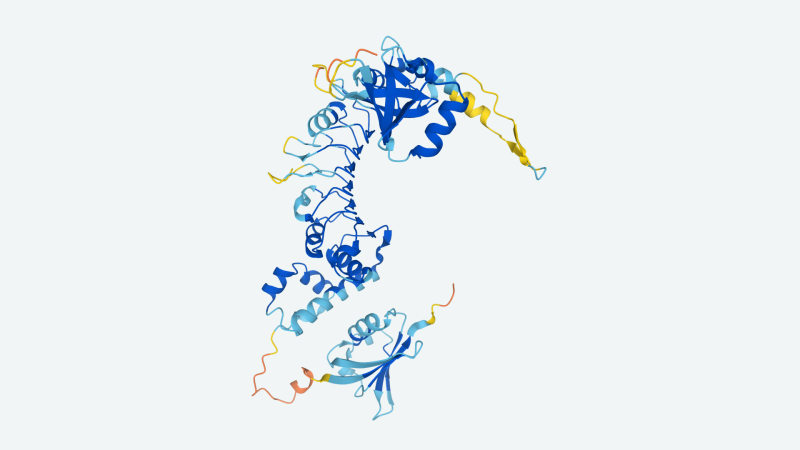
Image Credit: DeepMind
DeepMind says it is committed to generating AlphaFold 2 accessible “at scale” and collaborating with partners to discover new frontiers, like how numerous proteins kind complexes and interact with DNA, RNA, and compact molecules. Earlier this year, the organization announced a partnership with the Geneva-based Drugs for Neglected Diseases Initiative, a nonprofit pharmaceutical organization that hopes to use AlphaFold to determine compounds to treat circumstances for which drugs stay elusive. The Centre for Enzyme Innovation is applying the method to support engineer quicker enzymes for recycling polluting single-use plastics. And teams at the University of Colorado Boulder and the University of California, San Francisco are studying antibiotic resistance and SARS-CoV-2 biology with AlphaFold 2.
“Proteins are like tiny exquisite biological machines. The same way that the structure of a machine tells you what it does, so the structure of a protein helps us understand its function. Proteins are like tiny exquisite biological machines. The same way that the structure of a machine tells you what it does, so the structure of a protein helps us understand its function,” DeepMind CEO Demis Hassabis wrote in a weblog post published today. “At DeepMind, our thesis has always been that artificial intelligence can dramatically accelerate breakthroughs in many fields of science, and in turn advance humanity. We built AlphaFold and the AlphaFold Protein Structure Database to support and elevate the efforts of scientists around the world in the important work they do. We believe AI has the potential to revolutionise how science is done in the 21st century, and we eagerly await the discoveries that AlphaFold might help the scientific community to unlock next.”

/cdn.vox-cdn.com/uploads/chorus_asset/file/23262657/VRG_Illo_STK001_B_Sala_Hacker.jpg)
/cdn.vox-cdn.com/uploads/chorus_asset/file/25417841/TLC_Q6_Lifestyle_Press_Image.jpg)
/cdn.vox-cdn.com/uploads/chorus_asset/file/25184512/111323_PlayStation_Portal_ADiBenedetto_0010.jpg)
/cdn.vox-cdn.com/uploads/chorus_asset/file/25418195/04242024_Hollywood_Con_Queen_Post_big_image_post.jpg.large_2x.jpg)
